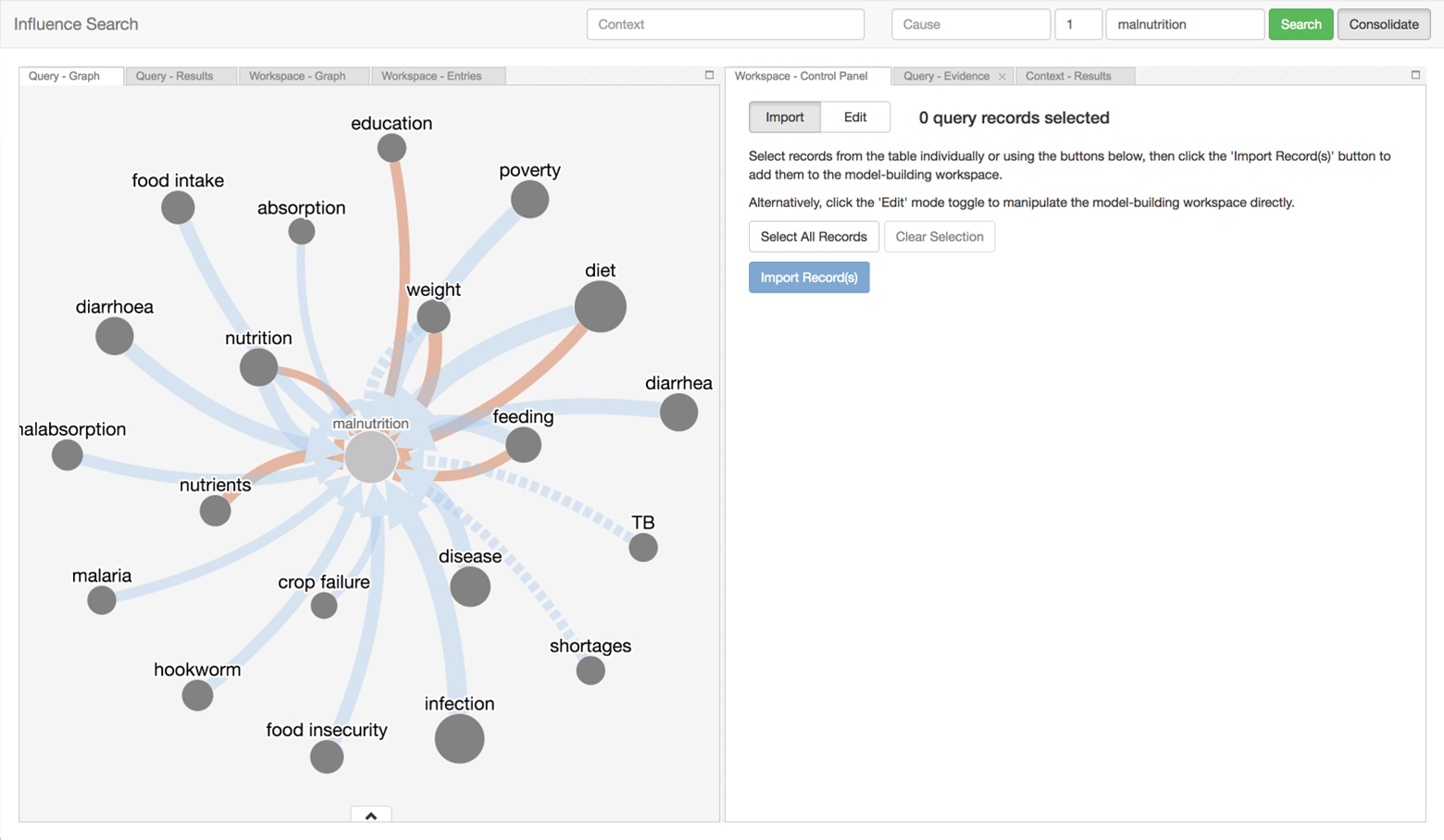
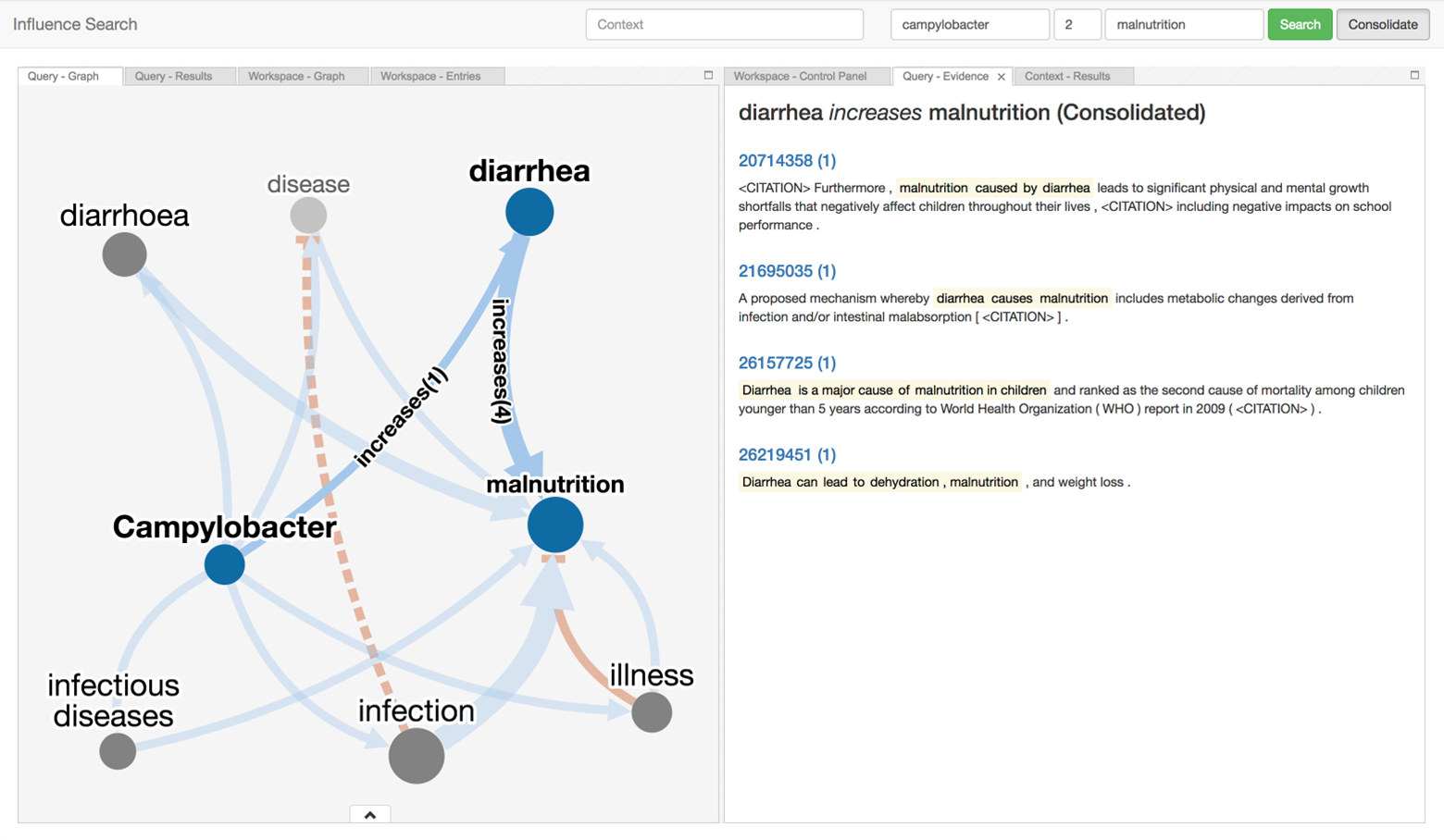
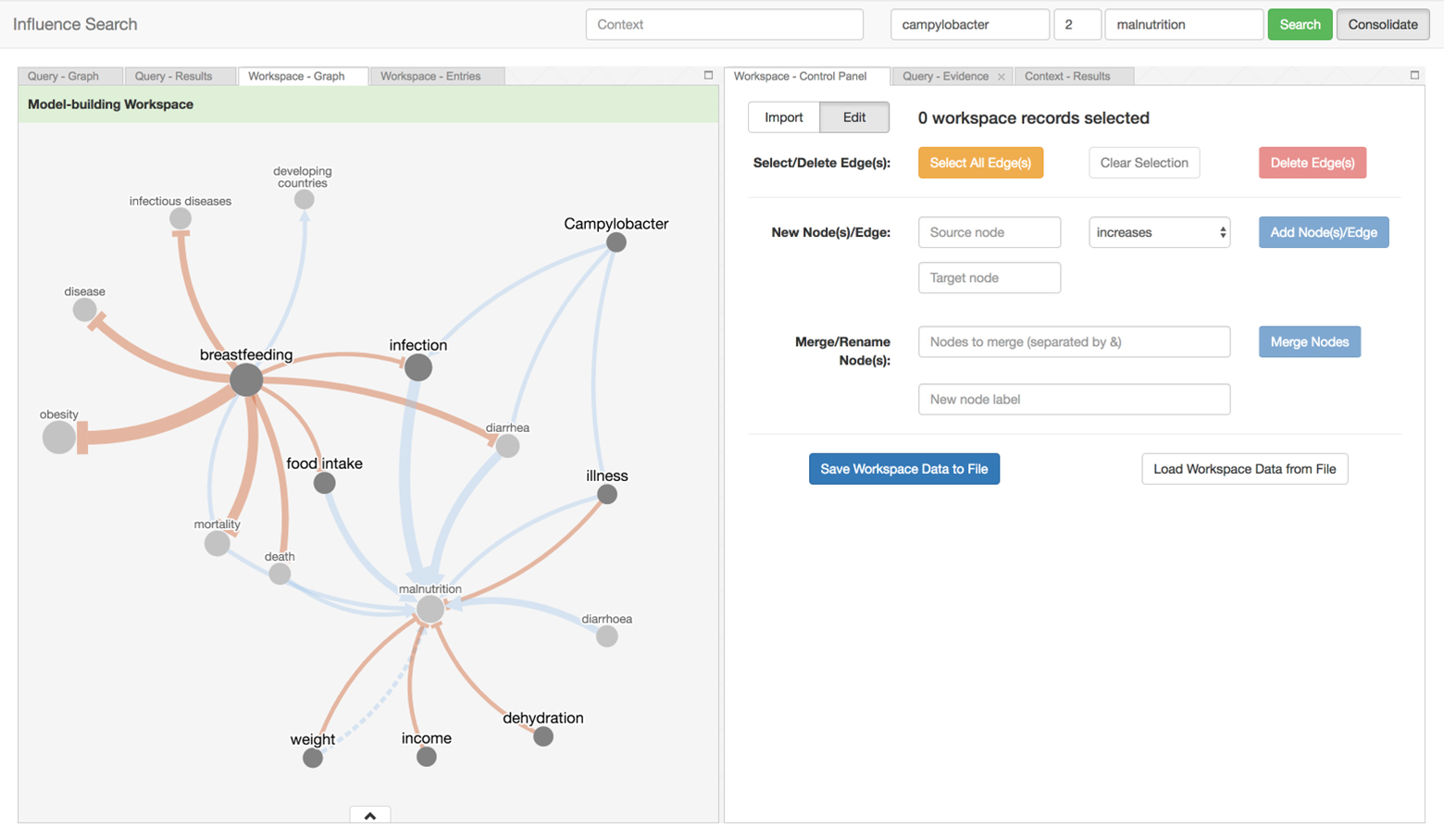
There is a vast amount of scientific findings scattered across the literature. Too many insights from research have gone undiscovered because fragments of independent but related knowledge have not been systematically retrieved, brought together, and interpreted. Influence Search provides a way to navigate the sea of research and assemble these puzzle pieces to help Ki understand both the scope of existing discoveries, and identify gaps in research that still need to be addressed.
Intended use
This tool is intended to be used by subject matter experts to explore and construct influence models in the domain of child growth and development. A second intended use is for decision-makers to understand opportunities for future investments to improve children’s health, both generally and in ways particular to different geographic settings.
Temporal timelines and intervention pathways
The tool can be used to search for influence pathways and aggregate them to into influence models that address various aspects of child growth and development. Thirdly, a collaboration between subject matter experts and decision-makers led to this tool being leveraged to construct a conceptual model of risk factors and mechanisms connecting infant growth restriction to later obesity.
The three main benefits of Influence Search
Start Date
July, 2016
Stage of Development
Beta Testing
Team Members
Marco Antonio Valenzuela-Escárcega, Gus Hahn-Powell, Zechy Wong, Mihai Surdeanu