Case Study
Using data governance and harmonization to learn about human breast milk: The International Milk Composition (IMiC) Consortium
- Data Sharing
Human breast milk provides infants with vital nutrition and immune protection through various beneficial compounds. However, the variability in composition between mothers remains poorly understood. The International Milk Composition (IMiC) Consortium aims to close this knowledge gap compiling a comprehensive dataset of breast milk components for high-level analysis and future research. Utilizing a robust data governance process, Ki is curating clinical metadata to integrate with biological data from milk samples across studies in both high-income countries (HIC) and low-and-middle-income countries (LMIC). Once completed, this dataset will enable analysis partners to develop and validate models that can identify optimal interventions for global maternal-child health.
BUILDING A DATASET TO ANALYZE MILK COMPOSITION
Human breast milk is not just food; it’s a complex biofluid evolved over millions of years to meet infants’ needs. It provides nutrition and immune protection through various compounds, including macronutrients (carbohydrates, proteins, fats), micronutrients (vitamins, minerals), bioactives (immune cells, growth factors, oligosaccharides, metabolites), and microbiota (bacteria, fungi). Despite the understanding that breast milk composition varies widely due to maternal health, diet, and environmental factors, surprisingly little is known about the specific determinants and consequences of this variation. Comprehensive analysis of breast milk variation could lead to greater improvements in child growth trajectories.
The IMiC Consortium, established in 2020, brings together maternal-child health researchers, laboratory partners, and statistical experts to analyze human milk composition. Ki leads the effort to design and implement a harmonized approach for assembling milk sample data and clinical metadata from four diverse regions: Tanzania, Pakistan, Burkina Faso, and Canada. The goal is to create a robust data governance process and a comprehensive dataset adhering to the FAIR Guiding Principles (Findable, Accessible, Interoperable, Reusable). This collaboration aims to leverage the expertise of nutritionists and analysts, ensuring that all partners work synergistically to focus on critical scientific and public health questions about optimal milk composition. By integrating their knowledge and working together as meaningful partners, rather than competitors, the consortium is positioned to develop and validate advanced models for enhancing maternal-child health interventions across different regions.
THE CHALLENGES OF DATA GOVERNANCE
Bringing together large datasets for analysis involves navigating significant complexities in data governance. Data governance encompasses the people, policies, and processes designed to ensure the quality, security, and compliance of data. The most pressing challenge is managing the diverse group of stakeholders, including researchers and data partners, who each bring varied perspectives and priorities to the table. Ki’s ability to build consensus among these stakeholders is crucial for efficient data sharing and analysis.
Securing data sharing policies across different jurisdictions is equally essential and challenging. For this project, Ki has been instrumental in proposing policies that enable seamless integration of study data across Tanzania, Pakistan, Burkina Faso, and Canada. These policies are designed to address local regulations, ethical considerations, and the unique requirements of each study site, ensuring privacy and compliance while facilitating ongoing data sharing between partners. Acquiring access to the study data from Canada posed the greatest challenge, as each Canadian province requires a separate legal agreement. We supported the University of Manitoba, another IMiC partner, in coordinating these agreements that permit the use of study data under delegated access, which limits access to authorized users under specific usage guidelines. Executing legal agreements across multiple nationwide studies takes time, patience and expertise.
After establishing collaboration and policy alignment, Ki undertook the complex task of harmonizing clinical metadata from the four studies: one longitudinal observational study from a HIC and three randomized controlled trials (RCTs) from LMICs. This metadata includes a wide range of variables, including maternal demographics and health records, breastfeeding practices, maternal-child nutritional intake and infant growth outcomes. It also includes data on interventions tested in the RCTs, including dietary supplements (Vitamin B3 or fortified food) and/or an antibiotic (Azithromycin)–variables Ki understands very well. Since each study was designed differently and provided data in different formats, creating a harmonized dataset required Ki’s proficient expertise in data cleaning and curation. Ki collaborated with site managers to obtain clean versions of the raw data, simultaneously guiding them through the data cleaning process. By identifying incompatibilities, sharing insights and suggesting solutions, Ki set a standard for data management that site managers could follow while ensuring consistent and comparable results.
In addition to cleaning and harmonizing each study’s clinical metadata, Ki created a seamless data flow process to integrate the metadata with data from human milk samples. The Consortium created a centralized biorepository to obtain human milk samples from each field site, housed at the Manitoba Interdisciplinary Lactation Centre (MILC) at the University of Manitoba in Canada. Lab partners then analyze these samples for a list of targeted milk components, generating what is referred to as omics data. In this context, omics data includes the spectrum of peptides, proteins, lipids, and metabolites in each milk sample, providing a comprehensive snapshot of biologic function as it relates to overall maternal health. This integration begins to reveal correlations and opportunities for intervention, particularly when comparing between HICs and LMICs.
TYPES OF DATA
Clinical data, breast milk data, omics data, outputs from analysis partners
COUNTRIES
Canada (CHILD study), Tanzania (ELICIT study), Pakistan (VITAL study), Burkina Faso (MISAMEIII study)
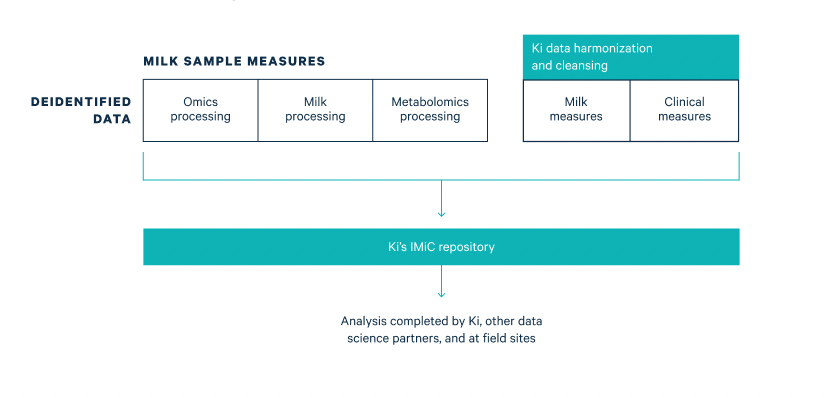
Milk sample measures, including omics and metabolomics data are cleaned, harmonized with clinical data, and stored in Ki’s repository for further analysis completed by Ki, other data science partners, and at field sites.
WORKING TOWARD TARGETED NUTRITIONAL INTERVENTIONS
Ki’s meticulous coordination of data cleaning, curation and harmonization, along with the creation of a robust data governance process, will help IMiC establish a comprehensive dataset on human milk composition. This ensures that all teams work with consistent data, enabling replicable results and allowing clinical sites, including those in LMICs, to access processed data from analytic partners in HICs. This dataset will provide unparalleled insights into the nutritional, bioactive and microbial components of breast milk across various geographic locations. By exploring how these components interact with each other and with nutritional supplements during lactation, our work will help the Gates Foundation and global teams better understand human milk composition, its complex relationships with maternal and infant factors, and optimize avenues for improving infant growth and other health outcomes.
Once completed, IMiC’s centralized database will enable analysis partners to apply statistical models that can identify optimal maternal, newborn and infant nutrition interventions. Complementary to its initial goals, IMiC and its data will influence other areas of intervention, including the safe and effective use of medications in breastfeeding mothers and their infants by enhancing pharmacokinetic (PK) models. These models, which predict how drugs are absorbed, distributed, metabolized, and excreted in the body, including their transfer into breast milk, provide crucial information for determining safe dosages. Understanding the variability in breast milk composition is essential for accurate predictions of drug transfer to infants that will inform optimal dosing strategies, particularly for infectious disease treatment and prevention in specific populations. Ki’s work with IMiC has been instrumental in laying the foundation for these advancements, supporting the work of countless collaborators to improve maternal-child health outcomes worldwide.
REFERENCES
- About IMiC. MILC. Accessed September 15, 2024. https://www.milcresearch.com/imic.html
- International Milk Composition (IMiC) Consortium. ClinicalTrials.Gov. Accessed September 15, 2024. https://clinicaltrials.gov/study/NCT05119166